Deep Learning-Based Morphological Classification between Classical Hodgkin Lymphoma and Anaplastic Large Cell Lymphoma: A Proof of Concept and Literature Review
Daniel Rivera, Kristine Ali, Rongzhen Zhang, Brenda Mai, Hanadi El Achi, Jacob Armstrong, Amer Wahed, and Andy Nguyen*
Department of Pathology and Laboratory Medicine, The University of Texas Health Science Center at Houston, Houston, TX, USA
*Corresponding author:Professor. Andy Nguyen, M.D., Department of Pathology and Laboratory Medicine, The University of Texas Health Science Center at Houston, 6431 Fannin, MSB 2.292, Houston, TX, 77030, USA, Telephone: (713) 500–5337, Fax: (713) 500–0712; Email: nghia.d.nguyen@uth.tmc.edu
Received: 24 October 2024; Revised: 11 November 2024Accepted: 12 November 2024; Published: 18 November 2024
Copyright: © 2024 Rivera D. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Citation: Rivera D, Ali K, Zhang R, Mai B, El Achi H, et al., (2024) Deep Learning-Based Morphological Classification between Classical Hodgkin Lymphoma and Anaplastic Large Cell Lymphoma: A Proof of Concept and Literature Review, 21st Century Pathology, Volume 4 (1): 159
Citation: Rivera D, Ali K, Zhang R, Mai B, El Achi H, et al., (2024) Deep Learning-Based Morphological Classification between Classical Hodgkin Lymphoma and Anaplastic Large Cell Lymphoma: A Proof of Concept and Literature Review, 21st Century Pathology, Volume 4 (1): 159
Abstract
Recent studies have shown promising results in using Deep Learning to detect malignancy in whole slide imaging. However, they were limited to predicting a positive or negative finding for a specific neoplasm. We attempted to use Deep Learning with a convolutional neural network algorithm to build a lymphoma diagnostic model specifically for difficult cases where two differential diagnoses are considered: classical Hodgkin lymphoma and anaplastic large-cell lymphoma. Our software was written in Python language. We obtained digital whole-slide images of hematoxylin and eosin-stained slides of 20 cases, including 10 cases for each diagnostic category. From each WSI, 60 image patches (100x100 pixels) at 20x magnification were obtained to yield 1200 image patches, from which 1079 (90%) were used for training, 108 (9%) for validation, and 120 (10%) for testing. For each test set of 5 images, the predicted diagnosis was combined from the prediction of five images. The test results showed excellent diagnostic accuracy at 100% for both image-by-image prediction and 100% for set-by-set prediction. This preliminary study provided a proof of concept for incorporating automated lymphoma diagnostic screens in specific scenarios into future pathology workflow to augment the pathologists’ productivity.
Keywords:
Deep learning, Classical Hodgkin lymphoma, Anaplastic large cell lymphoma
Introduction
Whole slide imaging (WSI) is increasingly used for research and education. Several efforts are being made to incorporate this technology into clinical work. A digital workflow requires additional equipment, such as an FDA-approved slide scanner, image storage, and a digital viewing workstation. Additionally, trained personnel and proper quality controls are needed. A pathology validation study conducted at Memorial Sloan Kettering Cancer Center to assess the pathologist's comfortability to render a primary diagnosis digitally in 2020 showed that 90% and 60% of pathologists were comfortable rendering a primary diagnosis digitally with and without access to glass slides, respectively. These numbers continue to increase as this technology becomes more broadly available to pathologists and trainees, reassuring the field about adopting new technologies. WSI systems have four main elements: a light source, a microscope with multiple lenses, a digital camera, and a system for repositioning the camera view along the tissue. The images can be scanned at different magnitudes, 20x and 40x being the most common. WSI scanners can use various modes, such as bright-field, fluorescent, and multispectral imaging [1-3].
Digital image analysis (DIA) offers features that can improve diagnostic accuracy and efficiency. On a digitized slide, histologic assessment using DIA demonstrates that cells can be classified by type and components, such as nuclei or cytoplasm. Moreover, areas of interest in the tissue can be mapped by shape or cell morphology and topology. The extraction of features from the tissue can be performed at different magnifications [4]. DIA still needs to overcome specific challenges; from a technical standpoint, it may be affected by different staining processes, as well as the fact that there is an unmet need to standardize the image acquisition process. Another aspect is the variation of tissue features in the setting of inflammation or malignancy, etc. Equally important is the biological variability of cellular and tissue features. Moreover, the size of the images, which rely on magnification and sensor pixel size, requires a significant amount of image storage [5]. Several platforms have been developed to overcome these challenges. These platforms can be divided into closed-source and open-source platforms, which can visualize, store, and analyze WSI. These platforms offer many processing, segmentation, and feature extraction tools with enough room to maximize their benefits [6-7].
Deep learning (DL) is a type of machine learning that encompasses different kinds of algorithmic formats such as convolutional neural network (CNN), recurrent neural network (RNN), long short-term memory (LSTM), and extreme learning model (ELM) [8]. DL has proven effective and reliable when applied in the medical field. The neuron is the fundamental unit; DL contains multiple layers of neurons, which allow high levels of abstraction and superior predictions from data input. Increasing the number of layers can enable more features to be detected and learned [9]. CNN is the algorithmic subtype that has gained recognition as the gold standard in image analysis with the adoption of parallel processing; as more efficient computing time has been achieved by performing all similar matrix operations simultaneously as opposed to linear sequences [10]. The core element of a CNN algorithm is convolution, which works by processing images using kernels (filters) to detect features of an image. The kernels move across the input to generate a feature map as a filtered image. The convolutional layers extract the features from an image patch to higher-level features, followed by the max-pooling layers to summarize data from the feature detection layer, reducing the computation in the network and leading to the result of predictions based on the given features [11].
The field of hematopathology has undergone significant evolution. The diagnostic process requires fine morphological skills in conjunction with precise interpretation of immunohistochemistry, flow cytometry, cytogenetics, and molecular testing. Integrating these elements to render a diagnosis of a hematolymphoid malignancy can be both challenging and stressful. Diagnosing a lymphoproliferative neoplasm will determine the course of action to be carried out by the clinicians, aiming for the best patient’s outcome. The global prevalence of all types of lymphoma is estimated to be about 545,000 cases each year [12]. These neoplasms encompass various conditions, ranging from indolent neoplasms to aggressive lymphomas. By histological assessment, several entities share morphological and/or architectural features, which can mislead the hematopathologist into the wrong diagnostic pathway, causing delayed or incorrect diagnosis and significantly impacting patient care. Another challenge is that hematopathologists are expected to render a diagnosis with smaller specimens, with high workloads due to the shortage of hematopathologists. Consequently, applying DL in clinical practice could improve efficiency, diagnostic precision, and the service's management. Our study aimed to develop a DL algorithm to differentiate between classical Hodgkin lymphoma (CHL) and anaplastic large-cell lymphoma (ALCL), both having complex morphologic features.
Materials and Methods
We conducted a retrospective compilation of cases with newly diagnosed CHL and ALCL by current World Health Organization criteria at our institution from 2017 to 2024 [13]. We reviewed the morphological characteristics of each case and selected the hematoxylin and eosin- (H&E) stained slides from 20 cases, which were scanned using the SG60 scanner (Philips Corporation, Amsterdam, Netherlands) at 40x magnification. The SG60 scanner has capacity for 60 glass slides, produces high-quality images, full automation (for focus, calibration, brightness and contrast settings), with tissue shape detection to outline and scan non-rectangular regions of interest for shorter turnaround times. The total scan time of a slide for a 15 × 15 mm benchmark scan area at a 40x resolution is ≤ 62 seconds. The images were acquired and stored in iSyntax2 format). Philips Image Management System was used to display the images. From each WSI, 60 image patches of 100x100 pixels (at 20x magnification, 0.5 μm/pixel) were obtained for feature extraction with SnagIt software (TechSmith Corp, Okemos, Michigan, USA). Each image patch is represented as a 100x100 matrix (100 rows and 100 columns) representing the intensity of 10,000 pixels. The image file was subsequently converted into a one-dimensional file with 30,001 entries; the first entry in the file stores the diagnostic label of the image, and entries from 2 to 30,001 store all the pixel intensity values (10,000 for each of the 3 color channels, Red-Green-Blue). A total of 1200 image patches were obtained from which 1079 (90%) were used for training, 108 (9%) for validation, and 120 (10%) for testing. The cases were divided into two cohorts, with 10 cases for each diagnostic category. For each test set of 5 images, the expected diagnosis was combined from the prediction of five images, i.e., at least three or more must agree to be considered as the predicted result (Figure 1). Our algorithm (with CNN model outlined in Figure 2) was written in Python using TensorFlow and Keras libraries [14, 15]. Parallel processing was performed using an NVIDIA GPU with compute unified device architecture (CUDA) [16]. The output was evaluated for diagnostic accuracy. This study was approved by the Institutional Review Board Ethics Committee per the Declaration of Helsinki.
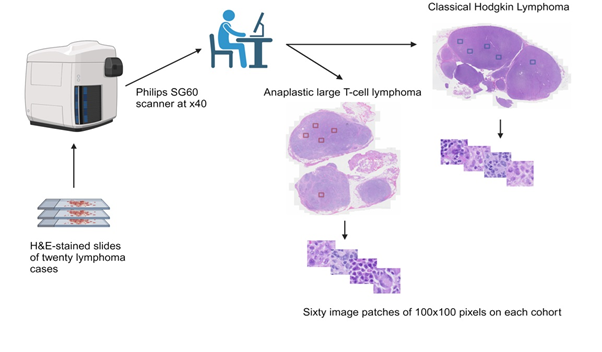
Figure 1: Case compilation, scanning, and feature extraction.
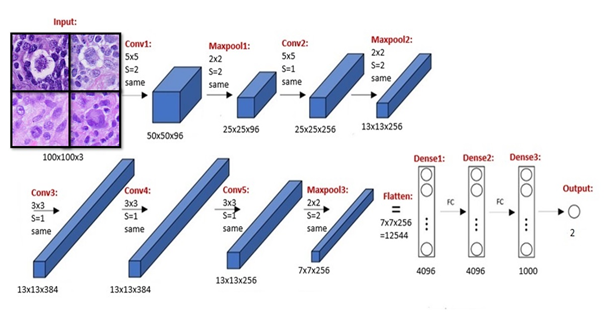
Figure 1: Algorithm written in Python in addition to TensorFlow and Keras libraries. Parallel processing was performed using NVIDIA GPU with compute unified device architecture (CUDA).
Results
Our model was trained and optimized for testing. It showed a diagnostic accuracy of 100% for both image-by-image and 100% for set-by-set prediction. Our preliminary study provided proof of concept for incorporating an automated lymphoma diagnostic screening model to help classify cases with challenging morphological features. We are confident these algorithms can be incorporated into future pathology workflows to augment hematopathologists’ productivity and diagnostic accuracy.
Discussion
Hematopathologists face challenges when diagnosing lymphoproliferative neoplasms due to overlapping morphological features that could lead to misclassification. Their role in patient care is critical; in the era of precision medicine. They are expected to be efficient, accurate and often have cases to diagnose with small specimens. CHL typically comprises large cells with large nuclei and prominent eosinophilic nucleoli, including the binucleated/multi-nucleated forms called Reed-Sternberg cells. The latter represent a minority of the neoplastic population. It is essential to render this diagnosis accurately because modern chemotherapy regimens achieve a cure rate of more than 80%. In contrast, ALCL is a mature T-cell neoplasm composed of large pleomorphic cells with abundant cytoplasm, and horseshoe/reniform nuclei with multiple nucleoli. This entity can mimic Reed-Sternberg cells and must be precisely distinguished because it can have more aggressive behavior and a lower cure rate than that of CHL [13]. Our study is the first to bring new insights into the applicability of a DL algorithm capable of performing morphological classification between CHL and ALCL, providing high diagnostic accuracy.
Over the years, several studies have shown the beneficial impact of implementing this technology in diagnosing lymphomas (Table 1). El Achi H, et al. (2019) (our DL research group) developed a DL model to differentiate between benign lymph nodes, diffuse large B-cell lymphoma (DLBCL), Burkitt lymphoma (BL), and small lymphocytic lymphoma (SLL). Four sets of 5 representative images, 40x40 pixels in dimension, were taken for each of 128 cases. A total of 2,560 images were obtained from which 1,856 were used for training, 464 for validation, and 240 for testing. The algorithm showed an accuracy of 95% for image-by-image prediction and 100% for set-by-set prediction [17]. Syrykh C, et al. (2020) evaluated the differences between follicular lymphoma (FL) and follicular hyperplasia (FH) on WSI scans from H&E-stained slides using a Bayesian neural network (BNN) and generated an overall accuracy of 91% with an area under the curve (AUC) of 0.99 [18]. Miyoshi H, et al. (2020) aimed to classify DLBCL, FL, and FH using DL on WSI scans. This classifier achieved the highest level of accuracy of 94%, 93%, and 92% for image patches with magnifications of x5, x20, and x40, respectively. They were also comparing the accuracy of the classifier to that of the pathologists. The accuracy of the classifier was 97%, whereas the pathologists achieved a lower accuracy of 83.3% [19]. Zhang J, et al. (2020) developed a model that included transfer learning using VGG-16 [20]. This method achieved an average five-fold cross-validation accuracy of 100%, 99.7%, and 99.2% to classify FL, chronic lymphocytic leukemia (CLL), and mantle cell lymphoma (MCL). Swiderska-Chadaj, et al. (2021) tested a DL model to predict the MYC translocation based on morphology on 287 cases of DLBCL from 11 hospitals; this algorithm achieved a sensitivity of 0.93 and specificity of 0.52; the authors concluded that their prediction model could save 34% of genetic testing [21]. Zhang X, et al. (2021) analyzed 374 WSI images, including CLL, FL, and MCL, and explored techniques to develop a diagnostic model based on recurrent neural network (RNN). The data was pre-processed using image flipping, color transformation, and other enhancement methods before inputting the data into the ResNet-50 network. The accuracy achieved with this algorithm was 98.63% [22]. Steinbuss G, et al. (2021) extracted images of WSI scans from H&E-stained slides from 629 patients to train and optimize an EfficientNet CNN algorithm, using Python libraries, with TensorFlow and R, on 84,139 image patches to classify SLL/CLL and DLBCL. The optimized model achieved an accuracy of 95.56% on an independent test set, including 16,960 image patches [23]. Yu WH, et al. (2021) developed a DL model to classify primary intestinal T-cell lymphomas and 40 WSIs were used to train the detection and segmentation of the nuclei of lymphocytes. The segmented cells were used to train a hybrid task cascade-region based convolutional network (HTC-RCNN) with ResNet50 as the backbone model. A decision tree-based machine learning (ML) algorithm, XGBoost, was trained to classify these lymphomas into two subtypes, including monomorphic epitheliotropic intestinal T-cell lymphoma (MEITL) versus intestinal T-cell lymphoma not otherwise specified (ITCL-NOS) to achieve an AUC of 0.966 [24]. Karabulut YY, et al. (2023) used a DL model to distinguish cases of mycosis fungoides (MF) versus non-MF cases by detecting the nuclei on WSI scans of H&E-slides. Ten nuclear properties were statistically significantly different between these two entities. Their lymphocyte detection model had an average prediction power of 90.5%, and the MF detection power algorithm showed an average prediction power of 94.2% [25]. Perry C, et al. (2023) trained a DL model using images of biopsies of aggressive B-cell lymphomas to screen and classify cases requiring FISH testing. This model showed a sensitivity of 100% and specificity of 87%, with an AUC of 0.95. This model presented a proof of concept of the applicability of this model as a screening tool when FISH analysis is limited [26]. Naji H, et al. (2024) proposed a HoVer-Net-base DL model for lymphoma segmentation to train a mask R-CNN model on WSI scans from H&E and immunohistochemistry (IHC) stained slides, achieving an F1 score of 0.899 and 0.913, respectively. The authors stated that their findings show potential improvement in lymphoma morphology and microenvironment assessment [27]. Lee JH, et al. (2024) worked on a prediction model with DINO (Self-distillation with no labels), a self-supervised architecture employing ViT-S/8, and multiple instances of learning (MIL) to evaluate pathological features in 216 cases of DLBCL status post-chemotherapy. The model yielded an AUC of 0.856, and these data highlight the potential applicability of the DL model as a diagnostic and prognostic tool for managing DLBCL [28]. Tagami M, et al. (2024) developed a screening DL model using PyTorch packages with pre-trained models and transfer learning to distinguish between IgG4-related ophthalmic disease (IgG4-ROD) and orbital mucosa-associated lymphoid tissue (MALT) lymphoma. Scans from H&E-stained slides were acquired, and the model after fivefold cross-validation achieved 73.3% accuracy with an AUC of 0.807 [29]. Zhang X, et al. (2024) developed and validated a DL model using LGNet for intraoperative diagnosis to distinguish primary central nervous system lymphoma (PCNSL) from other brain tumors by acquiring images from WSIs of frozen sections from different cohorts; this algorithm achieved an AUC of 0.981 and 0.993. The author concluded that the model outperforms some pathologists, and it does assist pathologists, irrespective of their experience, in improving diagnostic accuracy. Notably, the model also improves diagnostic accuracy in discriminating PCNSL from non-PCNSL, especially glioma (Table 1) [30].
Table 1: Deep learning experience for the diagnosis of lymphoma.
| Reference | Number of cases/images | Cohorts by disease | Algorithm format | Results |
|---|---|---|---|---|
| El Achi (PMID 31028058) | 128 cases | Benign LN DLBCL BL SLL |
CNN with TensorFlow and Keras libraries | 95% accuracy image by image 100% accuracy set by set |
| Syrykh (PMID 32377574) | 378 cases | FL FH |
CNN and BNN | 91% accuracy AUC 0.99 |
| Miyoshi (PMID 32472096) | 388 cases | DLBCL FL FH |
CNN | accuracy > 92% |
| Zhang J (PMID 32593219) | 374 images | CLL FL MCL | CNN with transfer learning using VGG-16, TensorFlow, and Keras libraries | accuracy > 99% |
| Swiderska-Chadaj (PMID 32979109) | 287 cases | DLBCL | CNN (U-net) | 93% sensitivity 52% specificity |
| Zhang X (PMID 33682770) | 374 images | CLL FL MCL |
RNN using ResNet-50 | 98.6% accuracy |
| Steinbuss G (PMID 34067726) | 629 cases | SLL/CLL DLBCL |
CNN using EfficientNet with R and TensorFlow libraries | 95.5% accuracy |
| Yu WH (PMID 34771625) | 40 cases | MEITL ITCL-NOS |
HTC-RCNN with ResNet50 XGBoost model |
AUC 0.966 |
| Karabulut (PMID 36571610) | 18 cases | MF non-MF |
CNN using sci-kit learn library | 94.2% prediction power |
| Perry (PMID 37958379) | 57 cases | Double/triple hit lymphomas | CNN adding MIL | 100% sensitivity 87% specificity AUC 0.95 |
| Naji (PMID 38237235) | 379 images | DLBCL | Mask R-CNN using HoLy-Net for segmentation | F1 score of 0.899 F1 score of 0.913 |
| Lee (PMID 38584594) | 216 cases | DLBCL | A model with DINO architecture employing ViT-S/8 and MIL | AUC 0.856 |
| Tagami (PMID 38700592) | 127 cases | IgG4-ROD Orbital MALT |
CNN using PyTorch packages with pre-trained models and transfer learning | 73.3% accuracy AUC 0.807 |
| Zhang X (PMID 38704409) | 1186 cases | PCNSL non-PCNSL |
CNN using LGNet | AUC 0.981 and 0.993 |
Despite the limited number of cases in our current study, the results provided a proof of concept using a deep learning algorithm with high diagnostic accuracy to distinguish 2 lymphomas with challenging morphological features. This model has demonstrated its potential to significantly impact the diagnosis of lymphomas. By leveraging its ability to learn complex patterns from large datasets of WSI, DL models can accurately classify lymphoma subtypes and could significantly assist hematopathologists. While progress has been made, several challenges and limitations remain to be addressed. Ensuring the quality and diversity of training datasets is crucial for model performance. Moreover, the interpretability (reasoning algorithm) of these models remains a challenge. Overcoming these obstacles will be essential for its broad adoption in clinical practice. DL offers a promising avenue for improving accuracy, efficiency, and consistency in the pathologist’s practice.
Conclusion
Despite the limited number of cases, our study demonstrates the effectiveness of a DL model in distinguishing complex cell morphology of classical Hodgkin lymphoma and anaplastic large-cell lymphoma, achieving 100% diagnostic accuracy. This high level of accuracy suggests that DL could serve as a powerful, supportive tool in lymphoma diagnosis, potentially reducing diagnostic variability and enhancing the efficiency of histopathological workflows.
There is a potential role for this DL model in clinical work as a QA tool. If the predicted diagnosis agrees with the histologic diagnosis by the pathologist, a final diagnosis can readily be confirmed. Otherwise, the case under consideration can be re-examined to ensure that no diagnostic features have been missed. In this way, the DL model serves as an extra checking step to help improving the diagnostic accuracy. Adopting DL models for lymphoma diagnosis could bring significant clinical benefits, particularly in challenging or ambiguous cases. DL-based systems could reduce diagnostic delays, improving patient outcomes. Furthermore, adopting DL in routine practice could alleviate the workload of hematopathologists. Integrating DL into diagnostic workflows might also enhance inter-laboratory consistency, especially in settings with limited access to specialized pathology expertise. Overall, this technology holds promise for improving the diagnostic accuracy and standardization of care in lymphoproliferative neoplasms.
Abbreviations
LN: Lymph node; DLBCL: Diffuse large B-cell lymphoma; BL: Burkitt lymphoma; SLL: Small lymphocytic lymphoma; FL: Follicular lymphoma; FH: Follicular hyperplasia; CLL: Chronic lymphocytic leukemia; MCL: Mantle cell lymphoma; MEITL: Monomorphic epitheliotropic intestinal T-cell lymphoma; ITCL-NOS: Intestinal T-cell, lymphoma, not otherwise specified; MF: Mycosis fungoides; IgG4-ROD: IgG4-related ophthalmic disease; Orbital MALT: Orbital mucosa-associated marginal zone lymphoma; PCNSL: Primary central nervous system lymphoma; CNN: Convolutional neural network; BNN: Bayesian neural network; HTC-RCNN: Hybrid task cascade- region based convolutional network; MIL: Multiple instance learning; HoLy-Net: HoVer-Net-based network; DINO: Self-distillation with no labels; AUC: Area under the curve.
Author Contributions
Daniel Rivera wrote the manuscript. Andy Nguyen mentored, reviewed, and edited the manuscript. All authors reviewed and approved the manuscript in its final form.
Conflict of Interest
The authors have no conflicts of interest to report.
References
- Hanna MG, Reuter VE, Ardon O, Kim D, Sirintrapun SJ, Schüffler PJ, et al. Validation of a digital pathology system, including remote review during the COVID-19 pandemic. Mod Pathol. 2020 Nov;33(11):2115-2127. https://doi.org/10.1038/s41379-020-0601-5
- Farahani N, Parwani A, Pantanowitz L. Whole slide imaging in pathology: advantages, limitations, and emerging perspectives. Pathology and Laboratory Medicine International. 2015 June;2015(7):23-33. https://doi.org/10.2147/PLMI.S59826
- Ghaznavi F, Evans A, Madabhushi A, Feldman M. Digital imaging in pathology: Whole-slide imaging and beyond. Annu Rev Pathol. 2013 Jan;8:331-359. https://doi.org/10.1146/annurev-pathol-011811-120902
- Escobar Díaz Guerrero R, Carvalho L, Bocklitz T, Popp J, Oliveira JL. Software tools and platforms in Digital Pathology: a review for clinicians and computer scientists. J Pathol Inform. 2022 June;13:100103. https://doi.org/10.1016/j.jpi.2022.100103
- Gutman DA, Khalilia M, Lee S, Nalisnik M, Mullen Z, Beezley J, et al. The digital slide archive: A software platform for management, integration, and analysis of histology for cancer research. Cancer Res. 2017 Nov;77(21):e75-e78. https://doi.org/10.1158/0008-5472.can-17-0629
- Wahab N, Miligy IM, Dodd K, Sahota H, Toss M, Lu W, et al. Semantic annotation for computational pathology: Multidisciplinary experience and best practice recommendations. J Pathol Clin Res. 2022 Mar;8(2):116-128. https://doi.org/10.1002/cjp2.256
- Humphries MP, Maxwell P, Salto-Tellez M. QuPath: The global impact of an open-source digital pathology system. Comput Struct Biotechnol J. 2021 Jan;19:852-859. https://doi.org/10.1016/j.csbj.2021.01.022
- Razzak MI, Naz S, Zaib A. (2018). Deep Learning for Medical Image Processing: Overview, Challenges and the Future. In: Dey N, Ashour A, Borra S. (eds) Classification in BioApps. Lecture Notes in Computational Vision and Biomechanics. Springer, Cham. 2018 Jan;26:323-350. https://doi.org/10.1007/978-3-319-65981-7_12
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015 May;521(7553):436-444. https://doi.org/10.1038/nature14539
- Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J Pathol Inform. 2016 Jul;7:29. https://doi.org/10.4103/2153-3539.186902
- Greenspan H, Van Ginneken B, Summers RM. Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique. IEEE Transactions on Medical Imaging. 2016 Apr 29;35(5):1153-9. https://doi.org/10.1109/TMI.2016.2553401
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021 May;71(3):209-249. https://doi.org/10.3322/caac.21660
- WHO Classification of Tumours Editorial Board. Haematolymphoid tumours. Lyon (France): International Agency for Research on Cancer. WHO classification of tumours series, 5th ed.; 2024;11. https://publications.iarc.who.int/637
- An end-to-end open-source machine learning platform. TensorFlow. https://www.tensorflow.org/
- Keras: The Python Deep Learning library. Available at: https://keras.io/
- Nvidia Cuda-Getting Started Guide for Microsoft Windows. DU-05349-001_v6.5, 2014 Aug.
- Achi HE, Belousova T, Chen L, Wahed A, Wang I, Hu Z, et al. Automated diagnosis of lymphoma with digital pathology images using deep learning. Ann Clin Lab Sci. 2019 Mar;49(2):153-160.
- Syrykh C, Abreu A, Amara N, Siegfried A, Maisongrosse V, Frenois FX, et al. Accurate diagnosis of lymphoma on whole-slide histopathology images using deep learning. NPJ Digit Med. 2020 May;3:63. https://doi.org/10.1038/s41746-020-0272-0
- Miyoshi H, Sato K, Kabeya Y, Yonezawa S, Nakano H, Takeuchi Y, et al. Deep learning shows the capability of high-level computer-aided diagnosis in malignant lymphoma. Lab Invest. 2020 Oct;100(10):1300-1310. https://doi.org/10.1038/s41374-020-0442-3
- Zhang J, Cui W, Guo X, Wang B, Wang Z. Classification of digital pathological images of non-Hodgkin's lymphoma subtypes based on the fusion of transfer learning and principal component analysis. Med Phys. 2020 Sep;47(9):4241-4253. https://doi.org/10.1002/mp.14357
- Swiderska-Chadaj Z, Hebeda KM, van den Brand M, Litjens G. Artificial intelligence to detect MYC translocation in slides of diffuse large B-cell lymphoma. Virchows Arch. 2021 Sep;479(3):617-621. https://doi.org/10.1007/s00428-020-02931-4
- Zhang X, Zhang K, Jiang M, Yang L. Research on the classification of lymphoma pathological images based on deep residual neural network. Technol Health Care. 2021;29(S1):335-344. https://doi.org/10.3233/thc-218031
- Steinbuss G, Kriegsmann M, Zgorzelski C, Brobeil A, Goeppert B, Dietrich S, et al. Deep Learning for the Classification of Non-Hodgkin Lymphoma on Histopathological Images. Cancers (Basel). 2021 May;13(10):2419. https://doi.org/10.3390/cancers13102419
- Yu WH, Li CH, Wang RC, Yeh CY, Chuang SS. Machine Learning Based on Morphological Features Enables Classification of Primary Intestinal T-Cell Lymphomas. Cancers (Basel). 2021 Oct;13(21):5463. https://doi.org/10.3390/cancers13215463
- Karabulut YY, Dinç U, Köse EÇ, Türsen Ü. Deep learning as a new tool in the diagnosis of mycosis fungoides. Arch Dermatol Res. 2023 Jul;315(5):1315-1322. https://doi.org/10.1007/s00403-022-02521-1
- Perry C, Greenberg O, Haberman S, Herskovitz N, Gazy I, Avinoam A, et al. Image-Based Deep Learning Detection of High-Grade B-Cell Lymphomas Directly from Hematoxylin and Eosin Images. Cancers (Basel). 2023 Oct;15(21):5205. https://doi.org/10.3390/cancers15215205
- Naji H, Sancere L, Simon A, Büttner R, Eich M-L, Lohneis P, et al. HoLy-Net: Segmentation of histological images of diffuse large B-cell lymphoma. Comput Biol Med. 2024 Mar;170:107978. https://doi.org/10.1016/j.compbiomed.2024.107978
- Lee JH, Song GY, Lee J, Kang S-R, Moon KM, Choi Y-D, Shen J, et al. Prediction of immunochemotherapy response for diffuse large B-cell lymphoma using artificial intelligence digital pathology. J Pathol Clin Res. 2024 May;10(3):e12370. https://doi.org/10.1002/2056-4538.12370
- Tagami M, Nishio M, Yoshikawa A, Misawa N, Sakai A, Haruna Y, et al. Artificial intelligence-based differential diagnosis of orbital MALT lymphoma and IgG4-related ophthalmic disease using hematoxylin-eosin images. Graefes Arch Clin Exp Ophthalmol. 2024 Oct;262(10):3355-3366. https://doi.org/10.1007/s00417-024-06501-1
- Zhang X, Zhao Z, Wang R, Chen H, Zheng X, Liu L, et al. A multicenter proof-of-concept study on deep learning-based intraoperative discrimination of primary central nervous system lymphoma. Nat Commun. 2024 May;15(1):3768. https://doi.org/10.1038/s41467-024-48171-x