Clinical significance of key genes in Pancreatic Cancer: Another Look
Raji Sundararajan*
School of Engineering Technology, Purdue University, West Lafayette, USA
*Corresponding author: Professor. Raji Sundararajan, School of Engineering Technology, Purdue University, West Lafayette, IN 47907-USA; E-mail: raji@purdue.edu
Received: 22 April 2023; Accepted: 17 May 2023; Published: 22 May 2023
Copyright: © 2023 Sundararajan R. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Sundararajan R (2023) Clinical Significance of Key Genes in Pancreatic Cancer: Another Look, 21st Century Pathology, Volume 3 (3): 148
Citation:
Abstract
The 94% death rate of pancreatic ductal adenocarcinoma indicates that current histopathological analyses and treatments do not adequately address the disease and thus in critical need of novel/additional treatment options for the different subsets of pancreatic cancer, the deadliest of all cancers. This is due to the inherent limitations of existing current practices. With its wide molecular heterogeneity and subsets, to analyze pancreatic ductal adenocarcinoma (PDAC), we need advanced technologies and know the clinicopathological characteristics of the genes and proteins in the tumors. Towards this, use of key genes/proteins as novel biomarkers and/or targets is an attractive alternative. Studying the various genes involved and their interactions offer a wealth of additional/novel information on the various genes, their pathways, and their impact on prognosis and treatment. The clinical relevance and significance of four key pancreatic cancer genes are analyzed and they are found to be potential targets for clinical practice.
Keywords:
Pancreatic cancer; Pancreatic ductal adenocarcinoma; SMAD4; p53
Introduction
With 466,000 deaths out of 496,000 cases (94%) worldwide, [1], pancreatic cancer needs urgent novel/additional techniques to diagnose and treat it. Both incidence and death rates are higher in high income countries, such as Europe, Northern America, Australia and New Zealand, due to obesity, diabetes, etc. Considering that the disease is either stable or slightly increasing, while breast cancer death rates are decreasing, it has been projected that pancreatic cancer will surpass breast cancer as the 3rd leading cause of cancer death by 2025 [1]. The 5-year survival rate is also only about 6% [2,3]. Pathology is one of the avenues to improve this, by using state-of-the art techniques, such as proteomics/genomics.
The difference between the normal and pathological, between a healthy and a malignant cancerous cell, arise from a variety of subtle causes [4]. It would seem reasonable to use these sensitivities and subtleties to characterize cancer tumors, and understand their basis, mechanisms that operate at the molecular and sub molecular levels. Using Genomics and Proteomics studies, it is possible to obtain the gene/protein profiles, and their interactions and pathways, and these techniques must be utilized to treat 21st century pancreatic cancers.
Molecular/protein analysis could improve outcomes with current treatments and accelerate therapeutic development through better biopsy techniques and analyses. Genomic and proteomic analysis could potentially enrich for therapeutic vulnerabilities and require proper clinical assessments. Study of protein-protein interactions of a cell/tissue is an attractive alternative to address the various subsets of pancreatic cancer. The current pathology methods do not address the various genomic subsets of the most common type (over 90%) of pancreatic ductal adenocarcinoma (PDAC) [5]. The mutational landscape of PDAC is highly heterogeneous [6, 7]. The diversity and complexity of somatic mutational processes underlying carcinogenesis could be revealed through mutational patterns observed in cancer proteomics and genomes. Elucidating underlying mutational processes using gene/protein studies will advance our standing of pancreatic cancer etiology with potential implications for better pathology and treatment. Towards this, the interactions of the four key genes of pancreatic are analyzed in this research for better outcomes to the patients.
Analysis of the four key genes
Table 1 shows the genetic profile of pancreatic carcinoma [8]. Out of these, the four key genes that are muted in most pancreatic cancers include Kirsten rat sarcoma viral oncogene (KRAS), tumor protein P53 (TP53), cyclin-dependent kinase inhibitor 2A (CDKN2A/p16), and Mothers against Decapentaplegic Homolog 4 (SMAD4) [3, 8, 9]. The frequency of their mutations is 95%, >90%, 75%, and 55% respectively [8,9]. Knowing the genes more frequently mutated in a cancer can have direct clinical impact; thus, it is of practical interest to study the interactions of these genes.
Table 1: Genetic Profile of Pancreatic Carcinoma [8].
|
Gene |
Gene locations |
% Frequency |
|
Oncogenes |
||
|
KRAS BRAF AKT2 GUCY2F NTRK3 EGFR EBV genome |
12p12.1 7q34 19q13.2
15q25.3 7p11.2 |
95 4 10-20 3 1 1 <1 |
|
Tumor suppressors/Genome-maintenance genes |
||
|
p16 TP53 SMAD4 BRCA2/PALB2 FANCC/FANCG MAP2K4 LKB1/STK11 ACVR1B TGFBR1 MSI-/TGFBR2 MSI+/TGFBR2 ACVR2 BAX MLH1 FABW7/cyclin E deregulation |
9p 17p13.1 18q21.2 13q13.1/16p12.2 9q22.32/9p13.3 17p12 19p13.3 12q13.13 9q22.33 3p24.1 3p24.1 2q22.3-q23.1 19q13.33 3p22.2 4q |
>90 75 55 8 3 4 4 2 1 1 4 4 4 4 6 |
|
Tissue-maintenance genes |
|
|
|
PRSS1 |
7q34 |
<1 |
Figure 1 shows the interactions of these genes [10]. The number of lines indicate the strength of the intractions: the higher the number between proteins, the stronger the interactions. There are strong interactions between CDKN2A and the other 3 genes; the weakest link is between KRAS and P53, while the interaction is moderate between P53 and SMAD4. Table 2 shows list of gene ontology (GO) enrichment of biological process, molecular function, and cellular component. Table 3 shows the list of various pathways.
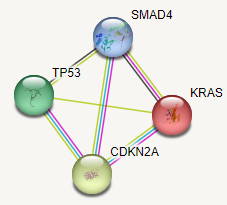
Figure 1: Interaction of the four pancreatic cancer proteins.
Table 2: List of GO enrichment.
|
Biological Process |
|
Positive regulation of cell aging Positive regulation of cellular senescence Replicative senescence Positive regulation of transcription from RNA polymerase Positive regulation of muscle cell apoptotic process |
|
Molecular Function |
|
MDM2/MDM4 family protein binding DISORDERED domain specific binding |
|
Subcellular Localization |
|
Beta-galactosidase complex chromatin |
Table 3: List of top 5 pathways.
|
KEGG Pathways |
|
Bladder cancer Pancreatic cancer Thyroid cancer Chronic myeloid leukemia Non-small cell lung cancer |
|
Reactome Pathways |
|
RUNX3 regulator CDKN1A transcription Oncogene induced senescence Transcriptional regulation by VENTX Transcriptional regulation by RUNX3 Oxidative stress induced senescence Genetic transcription pathway |
|
WikiPathways |
|
TP53 network Bladder cancer Extracellular vesicle-mediated signaling in recipient cells miRNA regulation of prostate cancer signaling pathways Head and neck squamous cell carcinoma |
Figure 2 shows the pancreatic cancer KEGG pathway [11]. The genetic alterations include KRAS and Her2 oncogenes and p16, p53, SMAD4, and BRCA2 tumor suppressor genes. Infiltrating ductal adenocarcinoma is the most common (90%) malignancy of the pancreas [9]. The diversity and complexity of various mutational processes underlying pancreatic cancer is many. The dominating genetic patterns are altered among the various diagnostic types of pancreatic carcinoma. The intraductal papillary mucinous neoplasms, the mucinous cystic neoplasms, acinar cell carcinomas, neuroendocrine carcinomas, pancreatoblastomas, and solid pseudopapillary neoplasms, however diverge significantly from the patterns of PanINs and typical invasive ductal carcinomas [9].
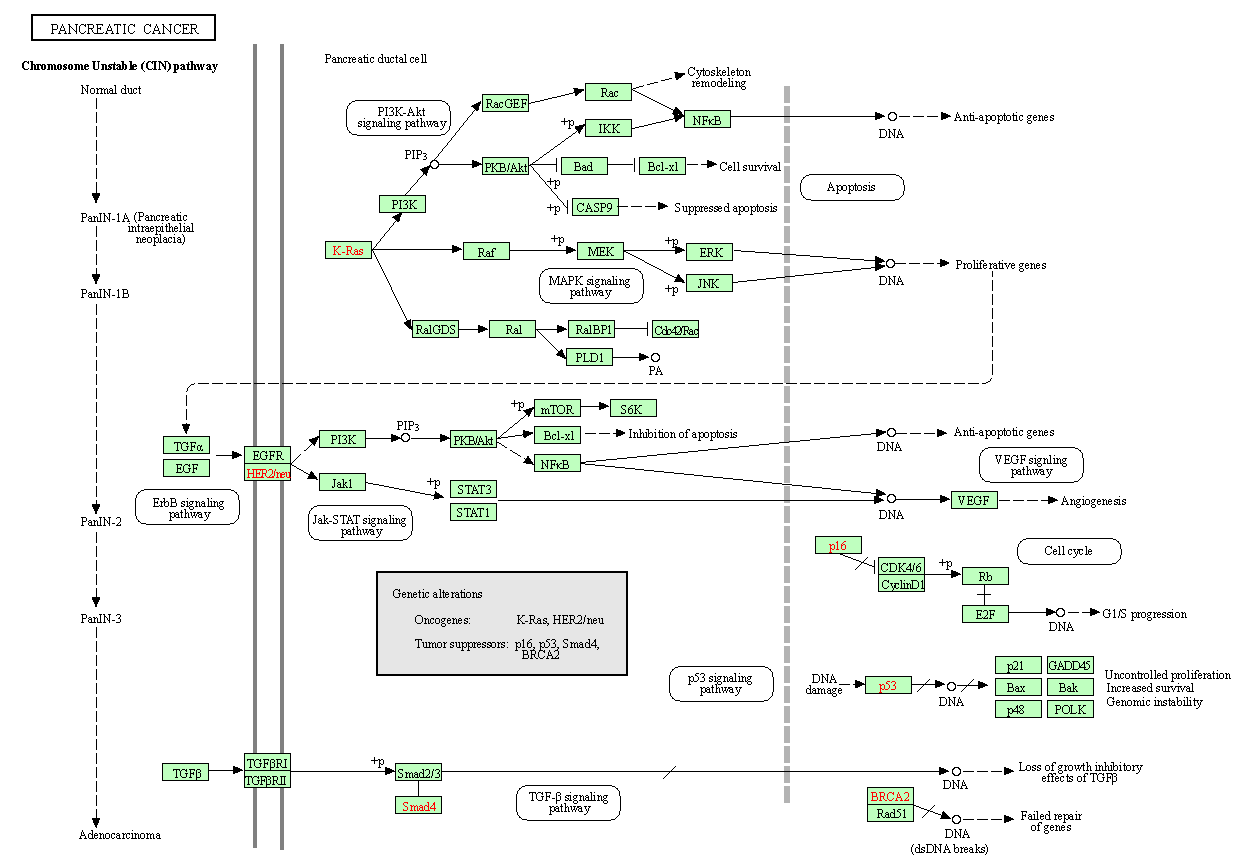
Figure 2: Pancreatic cancer KEGG pathway.
KRAS is a member of RAS family of genes that harbor mutations in various cancers. It is mutated in over 90% conventional PDAC. As one of the most commonly mutated genes in PDAC, KRAS is an attractive target for the development novel/additional therapies, and an understanding of the cancer biology of RAS protein could help in the development of the RAS-targeted therapies [8,3].
TP53 gene binds to specific sites of DNA and activates the transcription of various genes that control the cell division cycle and apoptosis. In approx. 75% of pancreatic cancers, TP53 gene has point mutations that inhibit the ability of p53 to bind DNA [8].
In general, many pancreatic cancers suffer a loss of p16 protein, through homozygous deletions or promoter methylation of the p16/CDKN2A gene associated with a lack of gene expression. In addition, inherited mutations of the p16/CDKN2A gene cause a familial melanoma/pancreatic cancer syndrome [8]. Patients with CDKN2A mutations tend to have poor overall survival [12].
Ductal adenocarcinoma is invasive, gland-forming, epithelial neoplasms [9]. Normal duct epithelium progresses to infiltrating cancer through a series of histologically defined precursor lesions, the pancreatic intraepithelial neoplasia (PanIN). The overexpression of HER-2/neu and activating point mutations in the KRAS gene occur early, while, inactivation of the p16 gene at an intermediate stage, and only at relatively late stages, the inactivation of p53, SMAD4, and BRCA2 occur. Activated KRAS engages multiple effector pathways. The various pathways include PI3-AKT signaling pathway, VEGF signaling pathway, p53 signaling pathway, and ErbB signaling pathway. PDAC shows extensive genomic instability and aneuploidy. Telomere attrition and mutations in p53 and BRCA2 are likely to contribute to these phenotypes. Inactivation of the SMAD4 tumor suppressor gene leads to loss of the inhibitory influence of the transforming growth factor-beta signaling pathway [8].
SMAD pathway mediates signals initiated on the binding of the extracellular proteins transforming growth factor-? (TGF?) and activin to their receptors. These signals are transmitted to the nucleus by the SMAD family of genes, including SMAD4. SMAD protein complexes bind specific recognition sites on DNA and cause transcription of various genes [8]. The SMAD4 gene is frequently (55%) mutated [8, 12]. Mutations in SMAD4 include both homozygous deletions and intragenic. The role of SMAD4 in pancreatic cancer has been explored in several studies [2,8,13-15].
In the cytoplasm, SMAD4 mediates signals from a family of TGF? ligands and their transmembrane receptors through the phosphorylation of SMAD proteins. Wang et al reported that SMAD4 mutation renders pancreatic cancer resistance to radiotherapy through the promotion of autophagy [13]. This was achieved through the induction of ROS and increased level of radiation-induced autophagy in Panc 1 and MIA PaCa-2 cell lines. The role of SMAD4 in regulating autophagy and radiosensitivity could be used as a novel biomarker for this frequently mutated gene in PDAC, which may have significant implication in deciding the proper treatment regimen for PDAC patients. A Meta-analysis of 1762 patients from 14 studies by Shugang X, et al. (2016) using STATA 12.0, and pooled hazard ratios, with 95% confidence intervals, revealed that loss of SMAD4 expression was found to be significantly correlated with poor overall survival [2]. Figure 3 shows the cytoscape pancreatic pathway [16].
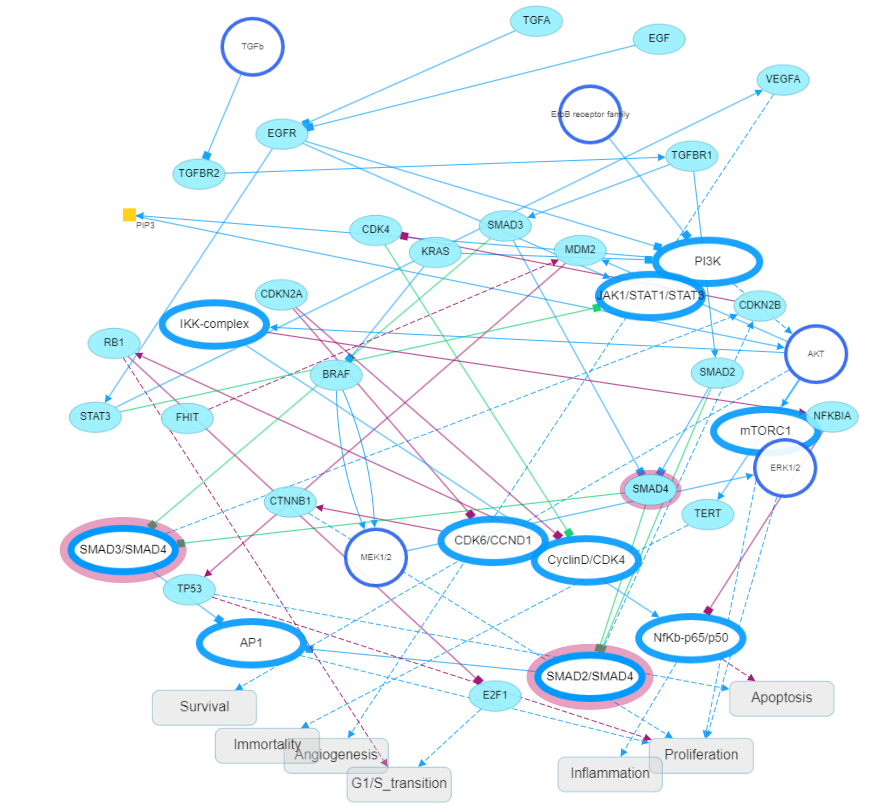
Figure 3: The cytoscape pancreatic pathway [16].
Conclusion
To treat pancreatic cancer effectively, we have to understand both the normal and pathological. The heterogeneous subtlety of biological processes of any cancer could well be explained by the various functions of genes/proteins. Without extending pathology to gene/protein dimension, the problem of cancer could not be solved, as indicated by the great number of pancreatic cancer deaths, each year worldwide. We cannot explain the subtle difference between a normal and a cancer cell without understanding the basic difference between gene/protein interactions and the various pathways.
The diversity and complexity of somatic mutational processes of the four key genes in pancreatic cancer could be studied more using molecular profiling and epidemiology. Collectively these studies will advance our standing of cancer etiology with potential implications for better pathology and treatment.
A better understanding of the key genes altered in pancreatic, such as SMAD4 may be a clinically useful biomarker for clinical responses. This could lead to a to novel gene-specific therapies for this deadly cancer.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2021 May;71(3):209-49. https://doi.org/10.3322/caac.21660
2. Shugang X, Hongfa Y, Jianpeng L, Xu Z, Jingqi F, Xiangxiang L, Wei L. Prognostic value of SMAD4 in pancreatic cancer: a meta-analysis. Translational oncology. 2016 Feb 1;9(1):1-7. https://doi.org/10.1016/j.tranon.2015.11.007
3. Dardare J, Witz A, Merlin JL, Gilson P, Harlé A. SMAD4 and the TGFβ pathway in patients with pancreatic ductal adenocarcinoma. International journal of molecular sciences. 2020 May 16;21(10):3534. https://doi.org/10.3390/ijms21103534
4. Schwarz G. R. Pethig: Dielectric and Electronic Properties of Biological Materials. John Wiley and Sons, New York, Chichester, Brisbane, Toronto 1979. 376 Seiten. https://doi.org/10.1002/bbpc.19800840137
5. Collisson EA, Bailey P, Chang DK, Biankin AV. Molecular subtypes of pancreatic cancer. Nature reviews Gastroenterology & hepatology. 2019 Apr;16(4):207-20. https://doi.org/10.1038/s41575-019-0109-y
6. Pompella L, Tirino G, Pappalardo A, Caterino M, Ventriglia A, Nacca V, Orditura M, Ciardiello F, De Vita F. Pancreatic cancer molecular classifications: from bulk genomics to single cell analysis. International Journal of Molecular Sciences. 2020 Apr 17;21(8):2814. https://doi.org/10.3390/ijms21082814
7. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, Boyault S. Signatures of mutational processes in human cancer. Nature. 2013 Aug 22;500(7463):415-21. https://doi.org/10.1038/nature12477
8. Scott E Kern and Ralph H Hruban. Pancreas Cancer. Ch 17, in Primer of the Molecular Biology of Cancer. Vincent T DeVita, Jr., Theodore S. Lawrence, Steven A. Rosenburg (Ed(s)), Wolters Kluwer, New York, 2011.
9. Sanchez D, Cheung WL. Pathology of pancreatic tumors. Translational Cancer Research. 2015 Dec 18;4(6):608-15. https://doi.org/10.3978/j.issn.2218-676X.2015.12.01
10. String-db.org. Accessed March 2023.
11. Pancreatic cancer - Homo sapiens (human). KEGG. https://www.kegg.jp/kegg-bin/show_pathway?hsa05212 accessed March 2023.
12. Principe DR, Underwood PW, Kumar S, Timbers KE, Koch RM, Trevino JG, Munshi HG, Rana A. Loss of SMAD4 is associated with poor tumor immunogenicity and reduced PD-L1 expression in pancreatic cancer. Frontiers in Oncology. 2022;12:154. https://doi.org/10.3389/fonc.2022.806963
13. Wang F, Xia X, Yang C, Shen J, Mai J, Kim HC, Kirui D, Kang YA, Fleming JB, Koay EJ, Mitra S. SMAD4 gene mutation renders pancreatic cancer resistance to radiotherapy through promotion of autophagy. Clinical Cancer Research. 2018 Jul 1;24(13):3176-85. https://doi.org/10.1158/1078-0432.CCR-17-3435
14. Sanh N, Fadul H, Hussein N, Lyn-Cook BD, Hammons G, Ramos-Cardona XE, Mohamed K, Mohammed SI. Proteomics profiling of pancreatic cancer and pancreatitis for biomarkers discovery. Journal of cell science & therapy. 2018;9(4). https://doi.org/10.4172/2157-7013.1000287
15. Wang C, Zhang T, Liao Q, Dai M, Guo J, Yang X, Tan W, Lin D, Wu C, Zhao Y. Metformin inhibits pancreatic cancer metastasis caused by SMAD4 deficiency and consequent HNF4G upregulation. Protein & cell. 2021 Feb;12(2):128-44. https://doi.org/10.1007/s13238-020-00760-4
16. cytoscape.org. Accessed March 2023.
